Molecular Systematics of the Genus Zoogloea and Emendation of the Genus
Molecular Systematics of the Genus Zoogloea and Emendation of the GenusYong Kook Shin, Akira Hiraishi, anf junta Sugiyama
Institute of Molecular and Cellular Biosciences, University of Tokyo, Yayoi 1-1 -1, Bunkyo-ku, Tokyo 113, and Laboratoy of Environmental Biotechnology, Konishi Co., Yokokawa, Sumida-ku, Tokyo 130, Japan Phylogenetic relationships among strains of zoogloea and related taxa were determined by 16s rDNA sequencing and genomic DNA hybridization techniques. The 16s rRNA gene was amplified by the polymerase chain reaction with a pair of eubacterial consensus primers and sequenced directly by using an automated fluorescent DNA sequencer. Sequence comparisons and distance matrix tree analysis revealed that Zoogloea ramigera IAM 12136 (= N. C. Dondero 106, type strain) and Zoogloea sp. ATCC 19324 formed a lineage with Rhodocyclus purpureus in the f3 subclass of Proteobacteria. 2. ramigera IAM 12670 (= P. R. Dugan 115) was shown to belong to another cluster with Akaligenes eutrophus and Pseudomonas cepacia in the B subclass. In contrast, Z. ramigera IAM 12669 (= K. Crabtree I-16-M) proved to be a member of the (Y subclass of the Proteobucteria, closely related to Agrobacterium tumefmiens. Genomic DNA hybridization studies also showed that there is genetic diversity among the strains currently designated 2. rarnigera, but typical Zoogloeu strains, characterized by their production of rhodoquinones, are highly related to each other and can be regarded as a single species. On the basis of the molecular data, together with the early phenotypic and chemotaxonomic information, we have emended the generic description of Zoogloea.
Strains of the genus Zoogloea Itzigsohn 1868, which now includes only one species, Zoogloea ramigera, are gramnegative aerobic chemoorganotrophic bacteria capable of forming the so-called zoogloeas, which are flocculent masses of cells embedded in gelatinous matrices and characterized bly a branched, finger-like morphology (8, 33). Since morphologically similar bacterial flocs often occur in biofilm and activated sludge systems, Zoogloea organisms are suggested to play an important role in wastewater purification processes.
Zoogloeal floc formation has been used for a long time as an important criterion for classification and identification of Zoogloea strains because of the lack of other distinct traits useful for this purpose. However, floc formation itself is found not only in Zoogloea strains but also in many other strains of metabolically similar bacteria of wastewater origin (3, 19, 21, 31). Unfortunately, the original isolate and other early isolates of Zoogloea (1, 7, 11, 22,34) have not been preserved and can no longer be compared in taxonomic studies. These situations have produced conflicting statements and much confusion as to the taxonomy and nomenclature of the genus Zoogloea, as reviewed by Zvirbulis and Hatt (38). In 1967, Crabtree and McCoy (2) proposed to designate strain I-16-M the neotype strain of 2. ramigera. A few years later, however, Unz (32), in a request for an opinion, proposed to reject strain I-16-M as the neotype strain and to replace it with strain 106, because the former strain was incapable of forming the true zoogloeas referred to by Itzigsohn (1868) in his original description of this species. The taxonomic confusion concerning the genus Zoogloea has begun to clarify since this proposal, but the current status of the genus is still unsatisfactory.
There exist three strains of 2. rarnigera that are wellknown through experimental use: strains 106T (T = type strain) (32), I-16-M (2), and 115 (8, 10). All these strains share the property of floc formation and were derived from wastewater environments, but marked phenotypic dissimilarities among them have been demonstrated. It has been suggested that the last two strains should be removed from the genus Zoogloea on the basis of phenotypic differences In a previous study, we used chemotaxonomic tools to provide a solution to the taxonomic problem concerning the genus Zoogloea (17). The most striking finding was that the typical strains of Zoogloea are characterized by their production of an unusual quinone, rhodoquinone-8 (RQ-s), in addition to ubiquinone-8 (Q-8), as a major quinone, whereas 2. ramigera I-16-M and 115 lack rhodoquinones. The chemotaxonomic data support the results of early phenotypic studies, improve the circumscription of the genus Zoogloea, and suggest the utility of rhodoquinone analysis as a new tool for identification of Zoogloea strains.
The genus Zoogloea has historically been considered a member of the family Pseudomonadaceae (25, 26), but this placement is tentative and based only on phenotypic information.
Phylogenetic relationships of Zoogloea strains to other pseudomonads have not yet been elucidated. The aim of the present study was to determine intra- and intergeneric relationships among strains of Zoogloea and related taxa by use of molecular techniques. The strategy taken involved polymerase chain reaction (PCR) amplification and direct automated sequencing of 16s rDNA, computer-assisted phylogenetic analysis, direct chromatographic analysis of DNA base composition, and colorimetric DNA-DNA hybridization assays. On the basis of the molecular data, together with previous phenotypic and chemotaxonomic information (16, 32, 33), we report here an emended generic description of Zoogloea.
Materials and methods
Bacterial strains and cultivation. Three strains of 2. ramigera, IAM 12136= (= strain 106T), IAM 12669 (= strain I-16-M), and IAM 12670 (= strain 115), and Zoogloea sp. strain ATCC 19324 were mainly used in this study. Zoogloea sp. strains ATCC 19173, ATCC 19123, AS180, AS456, and AS480 were also used in DNA homology studies. Some strains of established species of the p subclass of Proteobac teria (28) were added as reference organisms. For a list of test strains, see Table 2. Strains with IAM numbers were obtained from the Culture Collection Center of the Institute of Applied Microbiology, University of Tokyo (Tokyo, Japan); those with ATCC numbers were from the American Type Culture Collection (Rockville, Md.); and those with AS numbers are from our collection (17). A chemically defined medium designated LYS (17) was used for culturing Zoogloea strains, while a complex medium designated PBY (16) was used for all other organisms. Cells were grown aerobically in test tubes or Erlenmeyer flasks on reciprocal shakers at 30C, harvested by centrifugation at the early stationary phase of growth, washed with EDTA-saline (0.15 M EDTA plus 0.15 M NaC1, pH 8.5), and stored at -20C until used.
DNA extraction. Genomic DNA was extracted and purified by the method of Marmur (23). Since it was difficult to purify DNA from Zoogloea strains in the presence of the gelatinous matrix, the cells were treated with 0.1 N NaOH for 10 min at 4C to remove this matrix and washed twice with cold EDTA-saline prior to DNA extraction.
16s rDNA amplification. The 16s rRNA gene was amplified by PCR, for which 1 pg of genomic DNA was used as the template in a 100-pl reaction volume. The reaction was performed under a standard cycle condition by using a commercially available PCR reagent kit and a set of eubacterial consensus oligodeoxynucleotide primers as described previously (13). Amplified DNA was treated with chloroform, precipitated with ethanol, and purified by agarose gel electrophoresis, after which it was withdrawn with a Sephaglas BandPrep kit (Pharmacia LKB Biotechnology, Uppsala, Sweden) as specified by the manufacturer.
Sequencing. Five oligodeoxynucleotide primers labeled with fluorescein isothiocyanate at the 5 terminus were used for sequencing reactions. Four of the primers used corresponded to the complement of positions 1406 to 1389,1111 to 1093, 821 to 803, and 536 to 518 in Escherichia coli 16s rRNA numbering, as described previously (13). The remaining one was homologous to the sequence of positions 1091 to 1109 (5 -TAAGTCCCGCAACGAGCGC-3 ) of the E. coli numbering system. All these primers were obtained from Takara Suzo Co. (Kyoto, Japan). Sequencing reactions were performed by the linear PCR sequencing (cycle sequencing) method with the reagents from the Pharmacia AutoCycle sequencing kit, as described previously (13). The reaction mixtures were stored at -20C until analyzed, and immediately prior to electrophoresis, the mixtures were denatured by heating at 95C for 3 min and cooled quickly in ice-water.
Automated electrophoresis and analysis of DNA sequence reactions were performed by using a Pharmacia A.L.F.
DNA sequencer with 0.5-mm spacers and 5% acrylamide gels according to the manufacturers instructions.
Phylogenetic data analysis. Sequences were compiled from automatically and manually processed data of overlapping clones, aligned, and calculated for similarity by using the GENETYX program (Software Development Co., Tokyo, Japan) on an Apple Macintosh personal computer. Calculation of the nucleotide substitution rate (20) and construction of the distance matrix tree were performed by the neighborjoining method (27) with the CLUSTAL V program (12).
Alignment gaps and undetermined or ambiguous base positions were not taken into consideration for the calculations.
The bootstrap option, with 1,000 bootstrapped runs, in the CLUSTAL V program was also used to give the degree-ofconfidence intervals for each node on the phylogenetic tree.
DNA base composition and genomic DNA hybridization.
DNA base composition (mol% guanine [GI plus cytosine [C]) was determined by high-performance liquid chromatography of nuclease P1 hydrolysates of genomic DNA with external nucleotide standards (Yamasa Shoyu, Choshi, Japan) (14, 18). DNA-DNA homology studies were performed by the quantitative dot blot hybridization method with photobiotin labeling and colorimetric detection (9, 14).
Nucleotide sequence accession numbers. The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence data bases with the following accession numbers: 2. ramigera IAM 12136T, D14254; Z. ramigera IAM 12669, D14255; 2. ramigera IAM 12670, D14256; and Zoogloea sp. strain ATCC 19324, D14257. The accession numbers for sequences of organisms used as references are as follows: Agrobacterium tumefaciens, M11223; Alcaligenes eutrophus, M32021;
Alcaligenes faecalis, M22508; Alcaligenes xylosoxidans, M22509; Chromobacterium violaceum, M22510; Comamonas testosteroni, M11224; E. coli, 501859; Neisseria gonorrhoeae, X07714; Pseudomonas aeruginosa, M34133;
Pseudomonas cepacia , M22518; Pseudomonas diminuta, M59064; Rhodocyclus purpureus, M34132; Rochalimaea quintana, M11927; and Spirillum volutans, M34131.
Results
Sequence comparisons. The 16s rDNA fragments which corresponded to a nucleotide stretch from positions 7 to 1510 of the E. coli numbering system were amplified by PCR from four strains of Zoogloea and sequenced directly with a Pharmacia laser fluorescent DNA sequencer. The determined sequences covered approximately 95% of the entire 16s rRNA molecule. The number of determined sequences differed to some degree from strain to strain, ranging from 1,406 to 1,460 bases, because of the various deletions and additions of nucleotides found in these strains. The 16s rDNAs of Z. ramigera IAM 12136T and Zoogloea sp. strain ATCC 19324 were characterized by a deletion of 3 pairs in the loop at positions 77 to 92, an addition of G in the loop helix at positions 840 to 846, and a deletion at position 1140.
Z. ramigera IAM 12670 showed somewhat different features, including deletions of 1 base in the loop helix at positions 840 to 846 and 6 bases in the loop at positions 1131 to 1141. The 16s rDNA sequences of the above-mentioned three strains included the definitive signatures of the (3 subdivision of the class Proteobacteria reported by Woese (36). In contrast, Z. ramigera IAM 12669 16s rDNA was characterized by large deletions at positions 73 to 96, 200 to 217, and 452 to 479 and the addition of 2 bases at positions between 1010 and 1011, features that place this strain in the a subclass of the Proteobacteria.
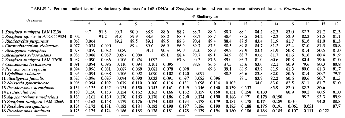
The sequences determined here were compared with a data set of 14 16s rRNA sequences from representative members of the class Proteobacteria, mainly those belonging to the (3 subclass. Computer-assisted pairwise sequence comparisons are shown in Table 1, where the upper right triangle gives overall percent similarity values and the lower left triangle shows the corrected values, which were calculated on the basis of unambiguously determined and alignable 1,250 positions of all sequences of the entire set.
Near-100% similarity was noted between Z. ramigera IAM 12136T and Zoogloea sp. strain ATCC 19324, both of which contain RQ-8 as a major quinone, as is the case for the typical Zoogloea strains (17). Among the organisms used for comparison, these Zoogloea strains were most similar to the phototrophic bacterium R. purpureus (91.3%). The typical Zoogloea strains had low similarities of less than 90% to the rhodoquinone-defective 2. ramigera IAM 12669 and IAM 12670. Relatively close similarities were found between 2. ramigera IAM 12670 and A. eutrophus (90.5%) and between 2. ramigera IAM 12669 and A. tumefaciens (95.2%).
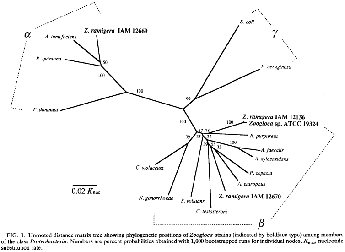
Phylogenetic tree analysis. A distance matrix tree was reconstructed by the neighbor-joining method from the data set of corrected distance values shown in Table 1. This phylogenetic tree is illustrated in Fig. 1. The topology of the tree has been evaluated by bootstrap analyses. The confidence values are given at branches which show possibly monophyletic fractions of related organisms separated at each node. The phylogenetic analysis showed that the typical strains of Zoogloea (IAM 12136T and ATCC 19324) formed a lineage with R. purpureus within the p subclass of Proteobacteria. The node placing these organisms as a monophyletic group was supported by 78% of the bootstrap confidence. 2. ramigera IAM 12670 was placed in a different lineage with A. eutrophus and P. cepacia within the p subclass, and this relationship is supported by 55% of the bootstrap replicates. In contrast, 2. ramigera IAM 12669 was deeply branched from the cluster encompassing members of the p subclass and was shown to be related to A. tumefaciens, a representative of the a subclass of Proteobacteria.
The node placing these organisms as a monophyletic group was supported by 50% confidence of the bootstrap analysis.
DNA base composition and DNA-DNA homology. The organisms used in this study included some Zoogloea strains for which the specific epithet has not yet been determined.
One of these strains, Zoogloea sp. strain ATCC 19324, has been shown to have a close phylogenetic relationship to 2. ramigera IAM 12136T by 16s rDNA sequence comparisons.
To know more about the intrageneric structure of Zoogloea strains, we studied DNA base composition of and DNADNA relatedness among the Zoogloea strains compared with some other related taxa.
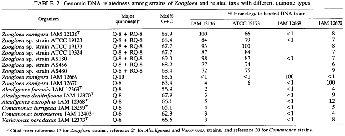
These results are shown in Table 2, in which quinone profiles of the test organisms are also presented for comparison.
Previous reports showed that the DNA base ratio of the type strain of 2. ramigera was 65.3 mol% as determined by buoyant density (32, 33). High-performance liquid chromatography experiments in the present study revealed that the G+C content of the DNAs of the RQ-8-producing Zoogloea strains, including 2. ramigera IAM 12136T, ranged from 67.7 to 69.0 mol%, being significantly higher than that previously reported for the type strain. The analytical method used here was a direct measurement of nucleotides derived from genomic DNAs and thus should provide more accurate data.
The rhodoquinone-defective 2. ramigera IAM 12669 and IAM 12670 showed much lower G+C contents than did the authentic Zoogloea strains. DNA-DNA hybridization assays showed that the RQ-&containing Zoogluea strains were highly related to each other, with reassociation values of more than 72%. As expected, there was no genetic relationship between the typical Zoogloea strains and the rhodoquinone- lacking Zoogloea strains or the reference organisms used for the p subclass of Proteobacteria.
Discussion
The results of the present study demonstrate the phylogenetic heterogeneity among the extant three strains of 2. ramigera and the phylogenetic unity between 2. ramigera IAM 12136T and other Zoogloea strains with rhodoquinones.
The 16s rDNA sequence-based phylogenetic analysis shows that the type strain of 2. ramigera and Zoogloea sp. strain ATCC 19324 form a lineage within the p subclass of Proteobacteria, with R. pulpureus as the closest relative. It is obvious that the genus name Zoogloea should be used for the limited number of strains belonging to this phylogenetic cluster. The phylogenetic tree positions 2. ramigera IAM 12670 in another branch within the p subclass, together with A. eutrophus and P. cepacia, the latter of which has been transferred into a new genus, Burkholderia, as Burkholderia cepacia (37). This fact suggests that 2. ramigera IAM 12670 should be removed from the genus Zoogloea and reclassified, possibly as a new genus and species relative to the above-noted two taxa. Since a strain the same as IAM 12670 has been and still is most widely used with the name 2. ramigera for experimental research in various fields (8), a nomenclatural proposal for this organism as a new taxon is imperative to end further taxonomic confusion. The deep branching of 2. ramigera IAM 12669 in the tree indicates that this Zoogloea organism is phylogenetically distant from the typical strains of Zoogloea and belongs to a lineage within the a subclass of the Proteobacteria. It is recommended that strain IAM 12669 should be reclassified as a known or new species of Agrobacterium or allied taxa.
The members of the family Pseudomonadaceae (25, 26) have proved to be so heterogeneous that they can no longer be united in one family. rRNA-DNA hybridization studies show that the genera Xanthomonas and Frateuria, representative genera of this family, have a distant phylogenetic relationship to Pseudomonas sensu strict0 within the y subclass of Proteobacteria (4-6). Apparently, the placement of the genus Zoogloea in the Pseudomonadaceae is inappropriate from phylogenetic standpoints.
Genomic DNA hybridization studies provide further evidence that there are close genetic relationships with homology values of more than 72% between the type strain of 2. ramigera and all test strains of Zoogloea for which the specific epithet has not been designated. In light of the genetic concept of a species standardized on the basis of DNA hybridization levels (35), we conclude that all these Zoogloea strains should be regarded as a single species, and at this time the genus Zoogloea is monotypic with only one species, 2. ramigera.
The genus Zoogloea has so far been defined and described on the basis of only phenotypic information in terms of morphology, physiology, and biochemistry (33). Included are active motility, floc formation, poly-P-hydroxybutyrate production, nitrate respiration, urease production, gelatin hydrolysis, and benzoate decomposition by meta cleavage of the ring structure. However, some of these characteristics seem too unreliable or nonspecific to be used for the definition of the genus. In concurrent phenotypic studies, we have found that active motility, poly-P-hydroxybutyrate production, gelatin hydrolysis, and urease production are different from strain to strain or depend on culture conditions. We prefer to remove these properties from the description of the genus Zoogloea. It has also been shown that nonflocculating variants of Zoogloea occur upon subculture (17, 29). Previous research has provided more valuable data on the circumscription of this genus from chemotaxonomic viewpoints (17). In particular, rhodoquinone production can be used as a distinct criterion for taxonomic purposes because the occurrence of this quinone structural type has been reported in quite a limited number of members among chemotrophic bacteria (15). The present report further gives the phylogenetic and genotypic information and revises the early data on DNA base composition. Thus, we emend the generic description of Zoogloea on the basis of the accumulated and revised information, together with some of the early phenotypic data (33), as follows.
Emendation of zoogloea Itzigsohn 1868. Straight to slightly curved rods, 1.0 to 1.3 pm in diameter and 2.1 to 3.6 pm in length. Motile by means of a single polar flagellum. Gram negative. Cells grown in liquid media form flocs or films which are embedded in gelatinous matrices and characterized by a finger-like or tree-like morphology. Nonflocculating variants sometimes occur. Growth occurs in mineral media supplemented with simple organic compounds as carbon sources and yeast extract as growth factor but is poor on ordinary nutrient agar containing peptone. Nonpigmented.
Aerobic chemoorganotrophs having a strictly respiratory type of metabolism with oxygen as the terminal electron acceptor. Anaerobic growth by nitrate respiration; denitrification positive. Oxidase positive. Weakly catalase positive. No acid is produced from glucose or many other carbohydrates. Benzoate is attacked by meta cleavage of the
ring structure. Major quinones are Q-8 and RQ-8. Palmitoleic acid (C16:l) is the major type of cellular fatty acid.
3-Hydroxy-decanoic acid (3-OH-C10:O) is present with smaller amounts of 3-OH-Cl2:O. The mol% G+C of DNA ranges from 67.3 to 69.0. Phylogenetic position is the P subclass of Proteobacteria . Habitat: polluted fresh water and biological wastewater treatment systems. Type species: Zoogloea ramigera Itzigsohn 1868.
1. Butterfield, C. T. 1935. Studies of sewage purification. 11. A zoogloea-forming organism found in activated sludge. Public Health Rep. 50:671-684.
2. Crabtree, K., and E. McCoy. 1967. Zoogloea ramigera Itzigsohn; identification and description. Int. J. Syst. Bacteriol. 17:l-10.
3. Deinema, M. H., and L. P. T. M. Zevenhuizen. 1971. Formation of cellulose fibrils by gram-negative bacteria and their role in bacterial flocculation. Arch. Mikrobiol. 78:42-57.
4. De Ley, J. 1992. The Proteobacteria: ribosomal RNA cistron similarities and bacterial taxonomy, p. 2111-2140. In A. Balow, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York.
5. De Vos, P., and J. De Ley. 1983. Intra- and intergeneric similarities of Pseudomonas and Xanthomonas ribosomal ribonucleic acid cistrons. Int. J. Syst. Bacteriol. 33:487-509.
6. De Vos, P., M. Goor, M. Gillis, and J. De Ley. 1985. Ribosomal ribonucleic acid cistron similarities of phytopathogenic Pseudornonas species. Int. J. Syst. Bacteriol. 35169-184.
7. Dias, F. F., and J. V. Bhat. 1964. Microbial ecology of activated sludge. I. Dominant bacteria. Appl. Microbiol. 12:412-417.
8. Dugan, P. R., D. L. Stoner, and H. M. Pickrum. 1992. The genus Zoogloea, p. 3952-3964. In A. Balow, H. G. Triiper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York.
9. Ezaki, T., S. Dejsirilert, H. Yamamoto, N. Takeuchi, S. Liu, and E. Yabuuchi. 1988. Simple and rapid genetic identification of Legonella species with photobiotin-labeled DNA. J. Gen. Appl. Microbiol. 34:191-199.
10, Friedman, B. A., and P. R. Dugan. 1968. Identification of Zoogloea species and the relationship to zoogloeal matrix and floc formation. J. Bacteriol. 951903-1909.
11. Heukelekian, H., and M. L. Littman. 1939. Carbon and nitrogen transformations in the purification of sewage by the activated sludge process. 11. Morphological and biochemical studies of zoogloeal organisms. Sewage Works J. 11:752-763.
12. Higgins, D. G., A. J. Bleasby, and R. Fuchs. 1992. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8:189-191.
13. Hiraishi, A. 1992. Direct automated sequencing of 16s rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15210-213.
14. Hiraishi, A., Y. Hoshino, and T. Satoh. 1991. Rhodoferax fernentans gen. nov., sp. nov., a phototrophic purple nonsulfur bacterium previously referred to as the Rhodocyclus gelatinosus- like group. Arch. Microbiol. 155330-336.
15. Hiraishi, A., and K. Komagata. 1989. Isolation of rhodoquinonecontaining chemoorganotrophic bacteria from activated sludge. FEMS Microbiol, Lett. 58:55-58.
16. Hiraishi, A., and K. Komagata. 1989. Effects of the growth medium composition on menaquinone homolog formation in Micrococcus luteus. J. Gen. Appl. Microbiol. 35311-318.
17. Hiraishi, A., Y. K. Shin, J. Sugiyama, and K. Komagata. 1992. Isoprenoid quinones and fatty acids of Zoogloea. Antonie van Leeuwenhoek 61:231-236.
18. Katayama-Fujimura, Y., Y. Komatsu, H. Kuraishi, and T. Kaneko. 1984. Estimation of DNA base composition by high performance liquid chromatography of its nuclease P1 hydrolysate. Agric. Biol. Chem. 48:3169-3172.
19. Kato, A., K. Izaki, and H. Takahashi. 1971. Floc-forming bacteria isolated from activated sludge. J. Gen. Appl. Microbiol. 12439.
20. Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120.
21. Kiuchi, K., H. Kuraishi, H. Murooka, K. Aida, and T. Uemura. 1968. Floc formation in activated sludge. 11. Identification of twelve representative strains isolated from activated sludge. J. Gen. Appl. Microbiol. 14:399409.
22. MacKinney, R. E., and M. P. Horwood. 1952. Fundamental approach to the activated sludge process. I. Floc-producing bacteria. Sewage Ind. Wastes 24:117-123.
23. Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:20&218.
24. Oyaizu-Masuchi, Y., and K. Komagata. 1988. Isolation of freeliving nitrogen fixing bacteria from the rhizosphere of rice. J. Gen. Appl. Microbiol. 34:127-164.
25. Palleroni, N. J. 1984. Family I. Pseudomonadaceae Winslow, Broadhurst, Buchanan, Krumwiede, Rogers and Smith 1917, 555AL, p. 141. In N. R. Krieg and J. G. Holt (ed.), Bergeys manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore.
26. Palleroni, N. J. 1992. Introduction to the family Pseudomonadaceae, p. 3071-3085. In A. Balow, H. G. Triiper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York.
27. Satiou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406425.
28. Stackebrandt, E., R G. E. Murray, and H. G. Triiper. 1992. Proteobacteria classis nov., a name for the phylogenetic taxon that includes the purple bacteria and their relatives. Int. J. Syst. Bacteriol. 38:321-325.
29. Strand, S. E., A. J. McDonnell, and R. F. Unz. 1988. Oxygen and nitrate reduction kinetics of a nonflocculating strain of Zoogloea ramigera. Antonie van Leeuwenhoek 54:245-255.
30. Tamaoka, J., D.-M. Ha, and K. Komagata. 1987. Reclassification of Pseudomonas acidovorans den Dooren de Jong 1926 and Pseudomonas testosteroni Marcus and Talalay 1956 as Comamonas acidovorans comb. nov. and Comamonas testosteroni comb, nov., with an emended description of the genus Comamonas. Int. J. Syst. Bacteriol. 3752-59.
31. Ueda, S., and R Earle. 1972. Microflora of activated sludge. J. Gen. Appl. Microbiol. 18:239-248.
32. Unz, R. F. 1971. Neotype strain of Zoogloea ramigera Itzigsohn. Int. J. Syst. Bacteriol. 21:91-99.
33. Unz, R. F. 1984. Genus IV. Zoogloea Itzigsohn 1868, 3PL, p. 214-219. In N. R. Krieg and J. G. Holt (ed.), Bergeys manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore.
34. Wattie, E. 1943. Cultural characteristics of zoogloea-forming bacteria isolated from activated sludge and trickling filters. Sewage Works J. 15476-489.
35. Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, 0. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Triiper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Tnt. J. Syst. Bacteriol. 37:463464.
36. Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:211-271.
37. Yabuuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group I1 to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251-1275.
38. Zvirbulis, E., and H. D. Hatt. 1967. Status of the generic name Zoogloea and its species. Int. J. Syst. Bacteriol. 17:ll-21.